Welcome to the Ogino MPE (Molecular Pathological Epidemiology) & EOC (Early-Onset Cancer) Lab Website
Better Health & Better World, through Integration of Molecular Pathology (Including Multi-Omics, Microbiology, & Immunology) and Population Health Science For Precision Medicine, Precision Prevention, and Precision Public Health.
Please also see the official page of Program in MPE Molecular Pathological Epidemiology in Department of Pathology, Brigham and Women’s Hospital.

Shuji Ogino, MD, PhD
Chief, Program in MPE Molecular Pathological Epidemiology, Department of Pathology, Brigham and Women’s Hospital
Professor of Pathology, Harvard Medical School
Professor in the Department of Epidemiology, Harvard T.H. Chan School of Public Health
Associate Member, Broad Institute of MIT and Harvard
Integrative Pathobiology-in-Population Sciences (IPPS): The 7th International MPE Meeting will be held in May 2025 in Buffalo, NY, USA
I am a transdisciplinary pathologist-epidemiologist in Harvard institutions, leading the Molecular Pathological Epidemiology (MPE) Program (in the BWH Pathology Department) and working in the interdisciplinary field of science-of-science. We integrate tumor microbiology/immunology, genomics, and microenvironmental analyses into population science frameworks to elucidate the biology/etiology of cancers including early-onset cancer. With my foresight, I have conceptualized several new methods, models, and paradigms with increasing relevance.
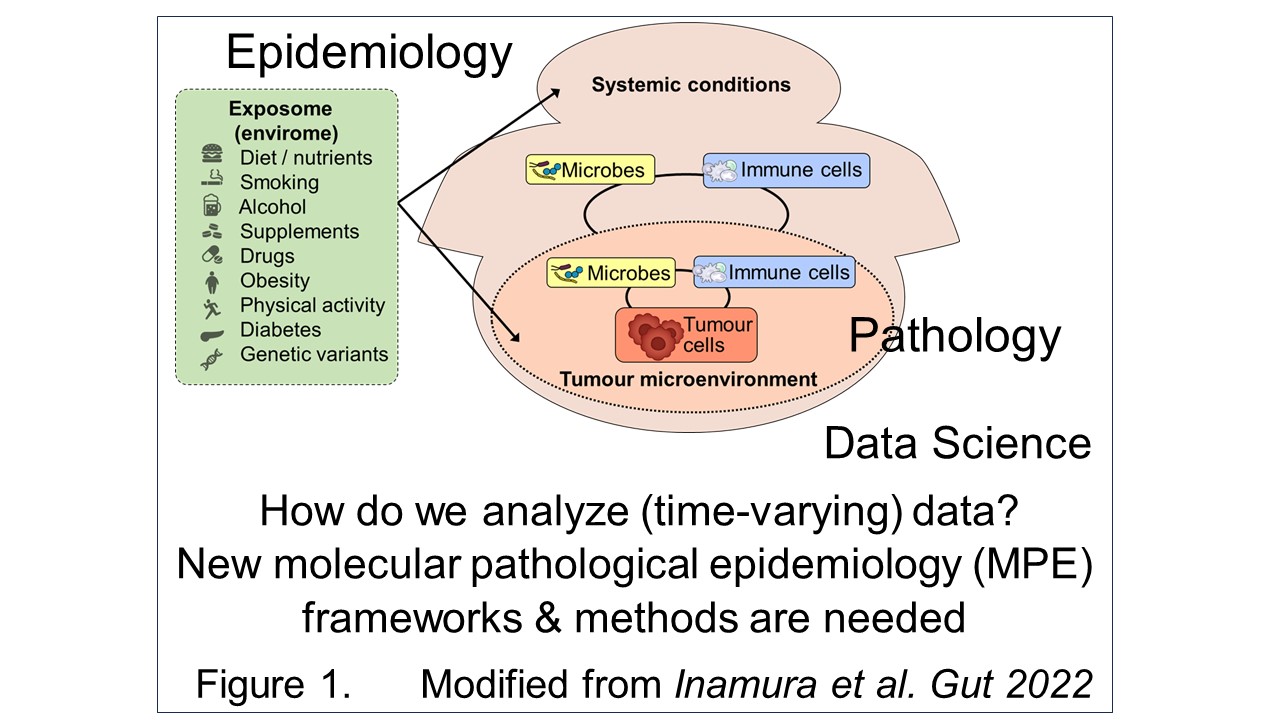
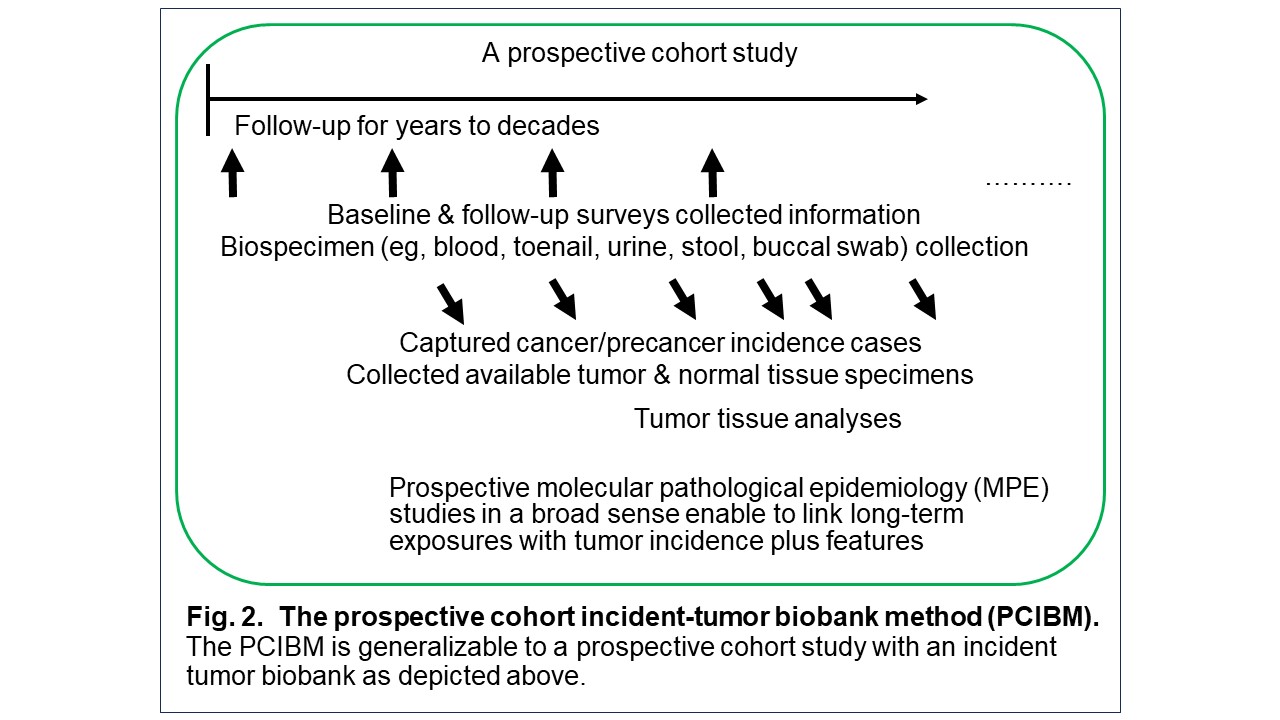
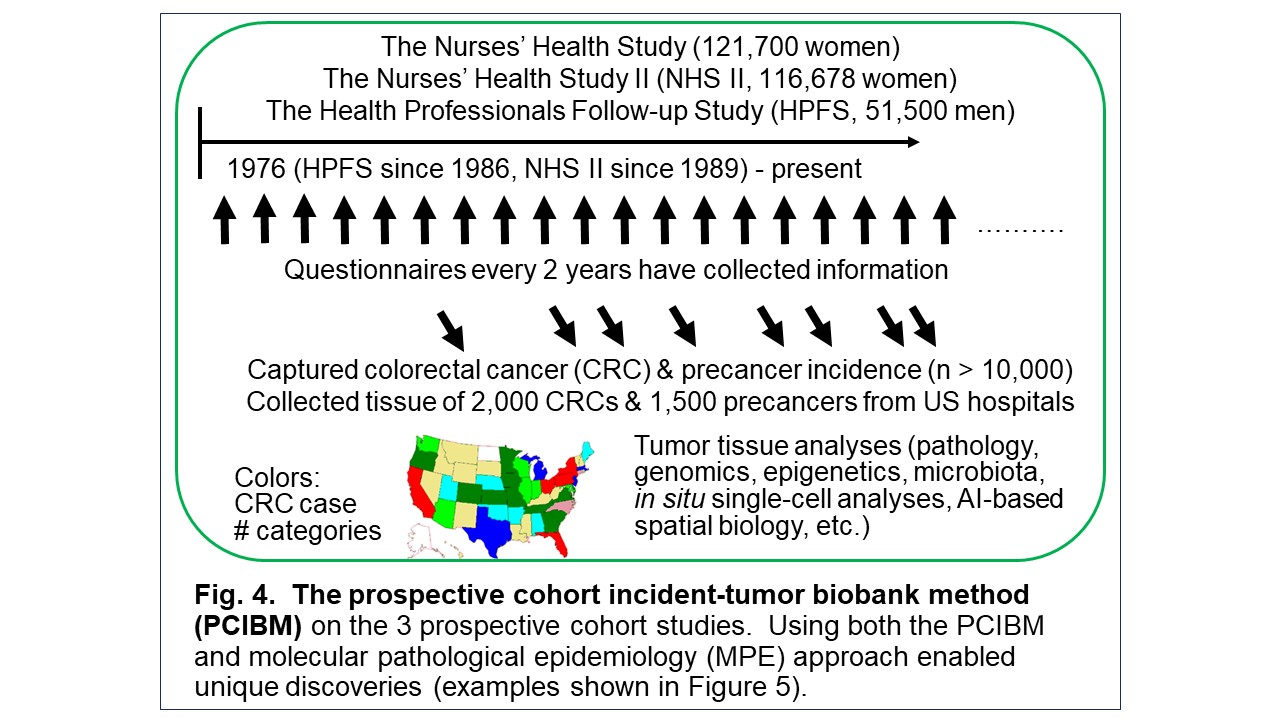
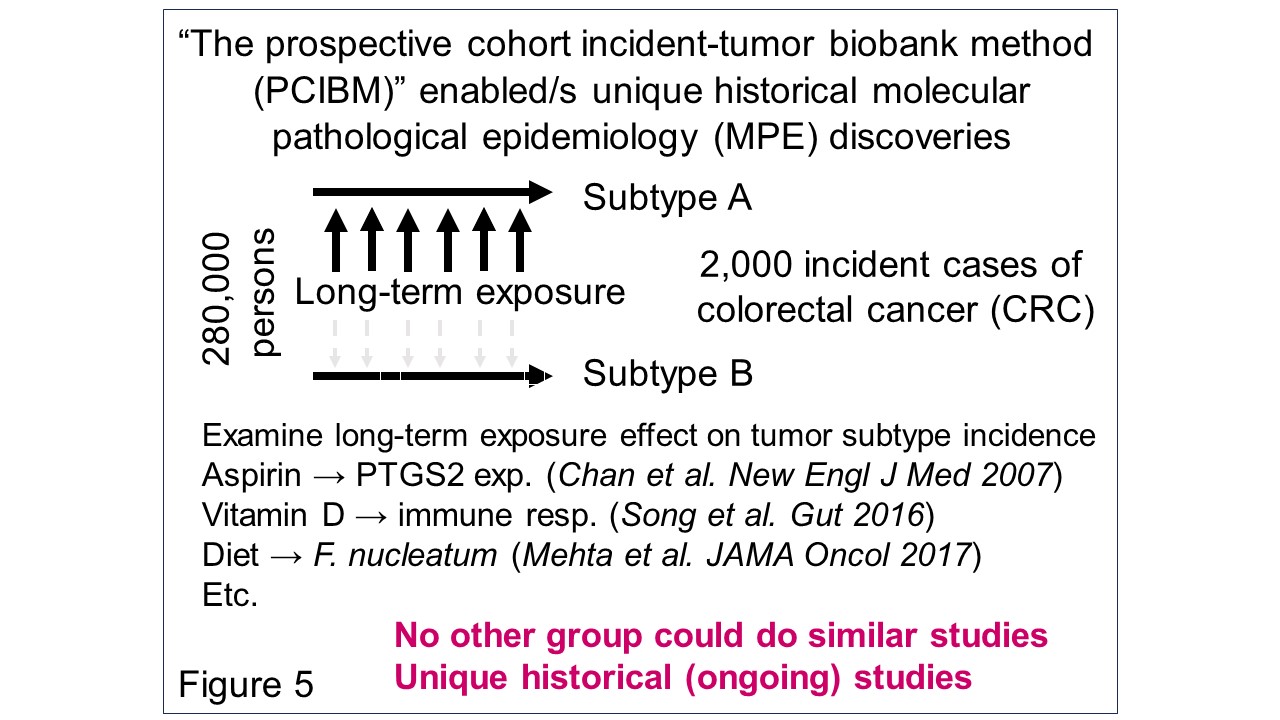
I have spearheaded the development of the interdisciplinary field of molecular pathological epidemiology (MPE) (Figure 1), widely recognized as one of the founders of the field. Tumor tissue collection efforts by many past/current investigators and staff members of the Nurses’ Health Study (NHS), NHS II, and Health Professionals Follow-up Study (HPFS) cohorts have led to a unique tissue biobank. We have conducted prospective MPE studies, using these unique resources and the prospective cohort incident-tumor biobank method (PCIBM). The PCIBM is an unconventional study method/design paradigm, which is generalizable to any prospective cohort with an incident tumor biobank (Figure 2). Using the three prospective cohorts, the PCIBM has enabled us to assess long-term exposures in relation to CRC incidence plus tumor characteristics, such as tumor pathology, microbial, molecular, immune, and microenvironmental features (Figures 3, 4). We have discovered many intriguing relationships of environmental, diet, and lifestyle factors with CRC incidence plus tumor molecular and microenvironmental features (Figure 5). Very few other groups have been able to conduct similar analyses. Hence, my lab has contributed substantially to a better understanding of the etiology and biology of CRC.
One of the most prominent features of myself is an unconventional innovative way of life. For example, please look at the keyboard on the right where keys are programmable and can be rearranged. I name this arrangement “smartly modified Dvorak in Kinesis”. Since 2000, I have been using it to increase my productivity. In the regular QWERTY key arrangement, most major keys are not conveniently placed. In my own special arrangement (“modified Dvorak”), my left fingers have “a – o – e – i (and u)” as home positions, and my right fingers have “(r and) h – t – n – s” as home positions; note these are the most commonly used letters. I thank Kinesis for manufacturing these types of keyboards!
This is a very efficient keyboard, but the only issue I have is that I have to carry this keyboard whenever I travel.
Narrative bio:
I am a pathologist and “molecular pathological epidemiologist” with a major research effort and 20% clinical effort in molecular diagnostics / molecular genetic pathology. Currently, I am the only unique faculty member who holds appointments in both Pathology and Epidemiology at Harvard University. I have played various major leadership roles in international and national societies. I have received prestigious awards and honors as listed below.
In 2010 (Ogino et al. JNCI 2010), I published to define the field of molecular pathological epidemiology (MPE) as a seamless integration of molecular pathology and epidemiology, which opened new opportunities. Until then, MPE-type analyses had been long recognized as part of molecular epidemiology (which is rather an ill-defined field). I described the basic concept and power of truly integrative science of MPE (Ogino et al. Gut 2011). I founded the International Molecular Pathological Epidemiology (MPE) Meeting Series in 2013 and have served as its Chair or Co-Chair. I continue to serve as a Co-Chair of its 6th meeting, which will be held in 2023. Locally, I founded the MPE Working Group in 2012 and continue to be its Leader. I have been developing new statistical framework and methods for MPE research in various settings (Wang et al. Am J Epidemiol 2015; Wang et al. Stat Med 2016; Nevo et al. Lifetime Data Anal 2018; Nevo et al. Biostatistics 2020; Li et al. Cancers 2022). In addition to the conceptualization of the unified MPE field, I have created several novel paradigms, concepts, and research frameworks, such as the GWAS-MPE approach (Ogino et al. Gut 2011), the colorectal continuum model (Yamauchi et al. Gut 2012), the unique disease principle (Ogino et al. Mod Pathol 2013), the etiologic field effect model (Lochhead et al. Mod Pathol 2015), the field of lifecourse MPE (Nishi et al. Am J Prev Med 2015), the field of social MPE (Nishi et al. Expert Rev Mol Diagn 2016), the field of pharmaco-MPE (Ogino et al. npj Precis Oncol 2017), the integrative field of immunology-MPE (Ogino et al. Lancet 2018; Ogino et al. Gut 2018), the causal inference MPE integration (Liu et al. Eur J Epidemiol 2018; Nevo et al. Int J Epidemiol 2021), and the field of microbiology-MPE (Hamada et al. J Pathol 2019; Mima et al. Hum Genet 2021) / microbiomics MPE (Inamura et al. Gut, in press). The MPE paradigm can transform and expand pathology (Ogino et al. Annu Rev Pathol 2019). Through these conceptual developments, I have proven the versatility of the MPE paradigm as well as the power of the interdisciplinary integration in one’s own mind (instead of a collaboration of separate experts). Productions of these new paradigms, frameworks, and research models are unique features of my scholarly activity. My MPE research using colorectal cancer as a disease model has shown intriguing links between risk factors and personal tumor characteristics, thereby refining effect estimate sizes of risk factor associations and supporting causality. Thus, our MPE research can contribute to the development of new strategies of precision cancer prevention and therapy.
With my unique combination of expertise in both molecular pathology and epidemiology, I have been teaching integration of molecular and population science at Harvard Medical School and affiliated hospitals, and Harvard T.H. Chan School of Public Health. I have served as a mentor for trainees with a wide variety of backgrounds in my interdisciplinary MPE laboratory, including pathologists, gastroenterologists, surgeons, epidemiologists, nutrition scientists, biostatisticians, and computational biologists. In addition, I regularly give a lecture on data science for pathology residents and fellows as a part of molecular diagnostics lecture series.
At Harvard T.H. Chan School of Public Health, I am a faculty member of Master of Science Program in Computational Biology and Quantitative Genetics (CBQ) directed by Dr. John Quackenbush ; Cancer Epidemiology and Cancer Prevention Track headed by Dr. Lorelei Mucci ; and Clinical Epidemiology
In summary, my research contribution has been very unique in integrating molecular pathology and population sciences as MPE, and has demonstrated widespread impact on biomedical and public health sciences.
List of Awards and Honors (Selected)
1999 Designee for CAP Foundation Scholar’s Award
1999 Resident Seminar Competition Finalists Award, from Ohio Society of Pathologists
2000 Pulmonary Pathology Society Pathologist-in-Training Award
2004 Executive Officer’s Award, from Association for Molecular Pathology (AMP)
2011 Ramzi Cotran Young Investigator Award, from United States and Canadian Academy of Pathology (USCAP)
2012 Meritorious Service Award, from Association for Molecular Pathology (AMP)
2014-present Elected Member, American Society for Clinical Investigation (ASCI)
2014 The Best of AACR Journals (selected as the corresponding author)
2014 “The Most Influential Scientific Minds: 2014″ by Thomson Reuters
2014-present Member, FASEB (Federation of American Societies for Experimental Biology) Excellence in Science Award Committee
2015 “The Most Influential Scientific Minds: 2015” and Highly Cited Researchers, by Thomson Reuters
2015-2022 NIH / NCI R35 Outstanding Investigator Award
2016 Highly Cited Researcher, by Thomson Reuters
2017 Highly Cited Researcher, by Clarivate Analytics
2018 Highly Cited Researcher, by Clarivate Analytics
2018 Runner-up for the Mentor-of-the-Year Award, Dana-Farber Cancer Institute
2018 Outstanding Investigator Award from American Society for Investigative Pathology (ASIP)
2019-present Member of OPTIMISTICC Team of Cancer Research UK’s Grand Challenge Award
2019 Highly Cited Researcher, by Clarivate Analytics
2019-2021 Invited nominator for Nobel Prize in Physiology or Medicine 2019, 2020, 2021
2020 Highly Cited Researcher by Clarivate Analytics
2021 Highly Cited Researcher by Clarivate Analytics
2022 Highly Cited Researcher by Clarivate Analytics
Education
(MD) University of Tokyo
(PhD in Pathology and Pathogenesis) University of Tokyo Graduate School of Medicine
(MS in Epidemiology) Harvard T.H. Chan School of Public Health
Hospital and academic appointments, and other roles
1994-1995 Intern (Japanese Physicians Medical Education Program), U.S. Naval Hospital Okinawa, Japan
1995-1997 Resident in Pathology, Allegheny General Hospital, Drexel University, Pittsburgh, PA (I met with and was tremendously influenced by Dr. Takanori Fukushima, who has been a renowned neurosurgeon with “god hands”
1997-1999 Resident in Pathology, Case Western Reserve University and University Hospitals Case Medical Center, Cleveland, OH (I did research in the lab of Dr. Sanford D Markowitz, and befriended fellow lab member Dr. William Grady)
1999-2001 Molecular Pathology Fellow (Fellowship Director, Dr. Debra Leonard) and Postdoctoral Fellow (in the lab of Dr. Robert B Wilson), University of Pennsylvania Medical Center, Philadelphia, PA
2001-2004 Instructor in Pathology, Harvard Medical School, Dana-Farber Cancer Institute, and Brigham and Women’s Hospital, Boston, MA
2001- Pathologist, Brigham and Women’s Hospital
2003- Member, Dana-Farber Harvard Cancer Center (Cancer Immunology Program, Cancer Epidemiology Program)
2004-2008 Assistant Professor of Pathology, Harvard Medical School, Dana-Farber Cancer Institute, and Brigham and Women’s Hospital
2008-2015 Associate Professor of Pathology, Harvard Medical School, Dana-Farber Cancer Institute, and Brigham and Women’s Hospital
2012-2015 Associate Professor, Department of Epidemiology, Harvard T.H. Chan School of Public Health
2013- Chair / Co-Chair, The International Molecular Pathological Epidemiology (MPE) Meeting Series
2015- Professor of Pathology, Harvard Medical School, and Brigham and Women’s Hospital
2015- Professor in the Department of Epidemiology, Harvard T.H. Chan School of Public Health
2016- Chief, Program in MPE Molecular Pathological Epidemiology, Department of Pathology, Brigham and Women’s Hospital
2017- Associate Member, Broad Institute of MIT and Harvard